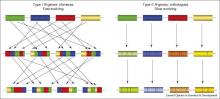
The majority of plant resistance (R)-genes encode a class of innate immune receptors (NLRs) with nucleotide binding and leucine-rich repeat 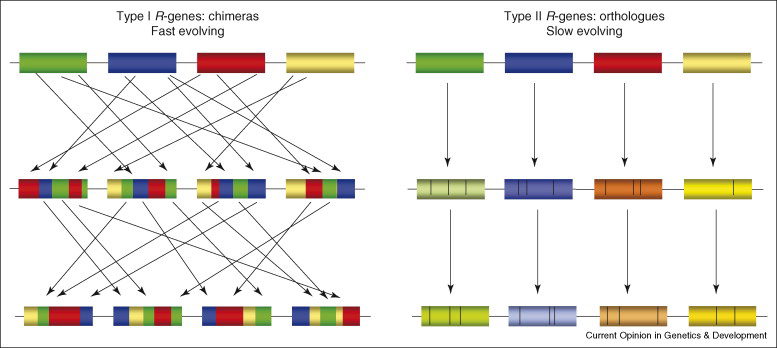 domains. Several NLRs act in multi-protein complexes that mediate interaction with pathogen effectors to induce resistance signaling. The complexity of these systems seems to have resulted from multiple rounds of plant–pathogen co-evolution. R-gene evolution is thought to be facilitated by the formation of R-gene clusters, which permit sequence exchanges via recombinatorial mispairing and generate high haplotypic diversity. This pattern of evolution may also generate diversity at other loci that contribute to the R-complex.
domains. Several NLRs act in multi-protein complexes that mediate interaction with pathogen effectors to induce resistance signaling. The complexity of these systems seems to have resulted from multiple rounds of plant–pathogen co-evolution. R-gene evolution is thought to be facilitated by the formation of R-gene clusters, which permit sequence exchanges via recombinatorial mispairing and generate high haplotypic diversity. This pattern of evolution may also generate diversity at other loci that contribute to the R-complex.
Genic and intergenic sequence repeats within R-clusters, generated by duplications and transposon insertions, provide a structural environment that permits mispairing during recombination, giving rise to unequal crossovers and interlocus gene conversions. Intergenic unequal crossover has the potential to place R-genes in new structural contexts that may alter expression, whereas intragenic mispairing generates chimeric genes that may encode novel functions. Both types of unequal recombination will also result in altered gene copy number within the cluster (gene duplication on one chromosome and loss on the other) according to the number of genes present in the region between the mispaired recombination sites.